Danthi Lab, Department of Biology, Indiana University

OVERVIEW
In the Danthi laboratory, we investigate the fascinating biology of dsRNA viruses. Our models of choice, mammalian reovirus and bacteriophage 6, infect hosts from two distinct domains of life but display many similarities both in how they are formed and in how they replicate. These viruses have multiple genome segments that are packaged within concentric protein shells. For each virus, after entry into cells, the genomic RNA remains within the particle. Instead, an intact inner shell is deposited into the cytoplasm. mRNA synthesized by a shell-enclosed RNA synthesis machinery then launches the viral replication program. Following synthesis of sufficient viral mRNA and proteins, the viral mRNA is packaged into newly formed virus assembly intermediates. The synthesis of viral dsRNA occurs within these progeny particles. Because of their unusual genome structure and replication strategy, these viruses have evolved novel strategies to successfully complete their replication cycle. Our goal is to uncover these mechanisms. We employ a variety of approaches including functional genomics, microscopy, cell and molecular biology and classical virology.
STUDIES ON MAMMALIAN REOVIRUS
Virus entry mechanisms. After attaching to host cell receptors, reovirus particles are internalized into cellular endosomes. Within cellular endosomes, the particles are disassembled and uncoated to release membrane targeting fragments that help deliver an intact core particle into the cytoplasm for replication. We are identifying and investigating factors that regulate virus disassembly and uncoating. We are also characterizing the interaction of viral components with host membranes that is important for cell entry.
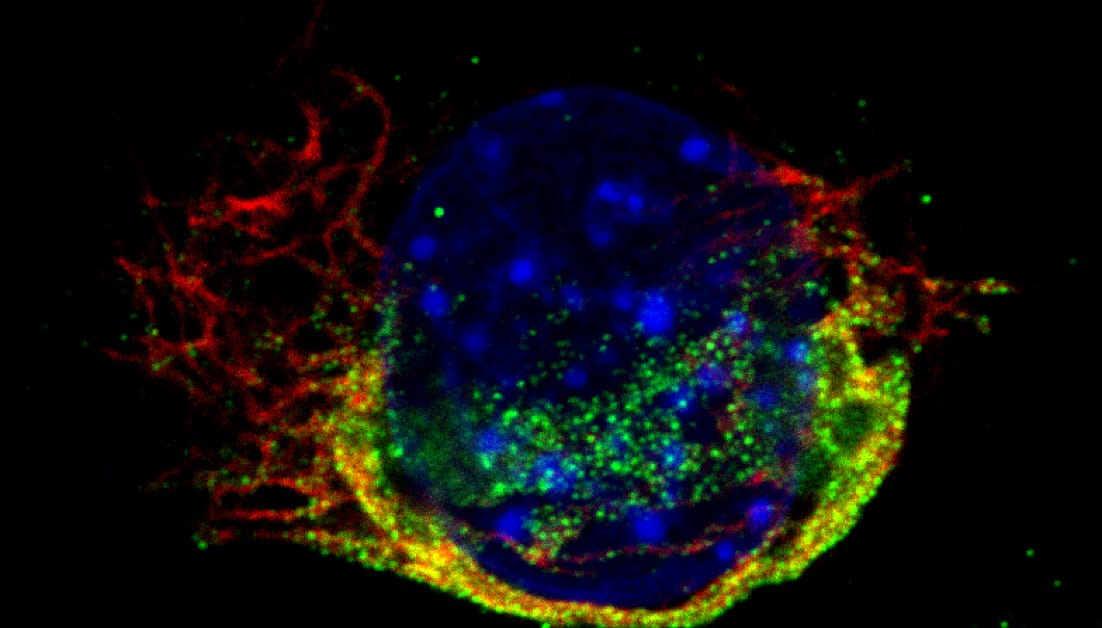
Host responses to virus infection. The reovirus genomic RNA is recognized by host cells to induce an antiviral innate immune response. We are investigating how reovirus genomic material that is encapsidated within concentric protein shells can become exposed for detection by host cells. We are also exploring the consequences of genomic RNA exposure to virus infection. Our results indicate that in addition to inducing the expression of cytokines such as interferon (IFN), reovirus RNA detection also results in induction of cell death. Our work is also focused on uncovering these pathways. Viruses that are sensitive to the activation of innate immune system during infection must evolve mechanisms to counter and limit this response. Our work has identified two different ways in which reovirus prevents full blown activation of this pathway. Some of our ongoing work is focused on delineating the underlying mechanisms.
STUDIES ON BACTERIOPHAGE PHI6
Some aspects of phi6 biology such genome replication, RNA packaging, capsid assembly and capsid structural features have been extensively studied for decades. The function of many phage proteins in these processes has been defined. In contrast, surprisingly little is known about the bacterial factors that are needed for phi6 replication. To identify such factors, we have completed a genome-wide transposon mutagenesis screen in its host organism, Pseudomonas syringae. We are currently working to uncovering Pseudomonas factors that are needed phi6 replication. We are particularly fascinated by how this phage crosses the two bacterial membranes during entry and exit. We are also interested in understanding if well-characterized phage defense systems can antagonize phi6 replication despite its unique features.